MFront
behaviour in pythonThis tutorial shows how we can use the Python MFrontGenericSupportInterface
to interact with a small-strain mechanical behaviour compiled with
MFront.
The corresponding behaviour is a small-strain von Mises plasticity
model with non-linear isotropic hardening and is implemented in the
SmallStrainPlasticity.mfront file using the StandardElastoViscoPlasticity
brick. Some material parameters (elastic parameters) are hard-coded
in the file while others (plastic parameters) are declared as material
properties to be specified by the user.
The following implementation shows:
External files
This tutorial can be run in a
jupyternotebook using the following files:
Regarding the final point, we aim to solve for a uniaxial stress state along the \(x\) direction, while controlling the imposed horizontal strain \(\epsilon\).
The remaining components \(\varepsilon_{ij}\) are considered as unknowns and we want to solve the following problem at a given imposed horizontal strain: find \(\boldsymbol{\varepsilon}\) such that \[ \boldsymbol{R}(\boldsymbol{\varepsilon}) = \begin{Bmatrix} \varepsilon_{xx} - \epsilon \\ \sigma_{yy}(\boldsymbol{\varepsilon}) \\ \sigma_{zz}(\boldsymbol{\varepsilon}) \\ \sigma_{xy}(\boldsymbol{\varepsilon}) \\ \sigma_{xz}(\boldsymbol{\varepsilon}) \\ \sigma_{yz}(\boldsymbol{\varepsilon}) \end{Bmatrix} = 0 \]
To solve this non-linear problem, we write a custom Newton-Raphson method which will involve the following jacobian: \[ \boldsymbol{J} = \dfrac{\partial \boldsymbol{R}}{\partial \boldsymbol{\varepsilon}} = \begin{bmatrix} \begin{bmatrix} 1 & 0 & \ldots & 0\end{bmatrix}\\ \bar{C}_\text{tang} \end{bmatrix} \] where \(\bar{\boldsymbol{C}}_\text{tang}\) is the sub-matrix obtained by removing the first row from the tangent stiffness \(\boldsymbol{C}_\text{tang} = \dfrac{\partial \boldsymbol{\sigma}}{\partial \boldsymbol{\varepsilon}}\).
In the following, we will compute the behaviour integration and the
above non-linear resolution for a batch of ngauss points
for illustration purposes. For simplicity, we will impose the same
strain \(\epsilon\) for all points,
yielding the same response for all points but different strain values
could also be considered for the different material points.
Note
To treat a batch of integration points, we rely on the
MaterialDataManagerclass. This is the most commonly used class inpython.In contrast, in solvers developped using a compiled language like
C++,fortranandC, most developers wants more control on how memory is handled. In this case,mgisprovides theBehaviourDataandBehaviourDataViewclasses which are suitable to deal with a single integraiton point.Those various solutions are described in depth in the
C++tutorial.
Note
This tutorial aims at illustrating how to use
MGISinpythonon a simple example.To describe a single integration point, the usage of the
mtestmodule (provided by theTFELproject if thepythonbindings are enabled) is strongly advocated.See this page for details.
First, we call the following command to compile the MFront behaviour:
mfront --obuild --interface=generic SmallStrainPlasticity.mfrontThis should only be done once and for all if the behaviour implementation does not change anymore.
Shared libraries are then stored in the
src/libBehaviour.so file which will be used to load the
behaviour.
import subprocess
import numpy as np
import mgis.behaviour as mgis_bv
behaviour_name = "SmallStrainPlasticity"
subprocess.call(["mfront", "--obuild", "--interface=generic", behaviour_name+".mfront"]);Treating target : all
The following library has been built :
- libBehaviour.so : SmallStrainPlasticity_AxisymmetricalGeneralisedPlaneStrain SmallStrainPlasticity_Axisymmetrical SmallStrainPlasticity_PlaneStrain SmallStrainPlasticity_GeneralisedPlaneStrain SmallStrainPlasticity_TridimensionalNote
If the
TFELproject (which providesMFront) has been compiled with thepythonbindings enabled, then themfrontpythonmodule provides a more direct way of compilingMFrontbehaviours, allowing to retrieve the name of the generated libraries and behaviours.See this page for details about the
mfrontmodule.
We first need to choose the modeling hypothesis. Here, we will work
with 3D states and thus choose the Tridimensional
hypothesis. Behaviours are loaded differently depending on whether we
work with a small or finite-strain setting. We use the
isStandardFiniteStrainBehaviour function to check whether
the behaviour is finite strain or not. The finite-strain case is
described in another demo.
lib_path = "src/libBehaviour.so"
hypothesis = mgis_bv.Hypothesis.Tridimensional
is_finite_strain = mgis_bv.isStandardFiniteStrainBehaviour(lib_path, behaviour_name)
if is_finite_strain:
print("Finite strain behaviour detected.")
bopts = mgis_bv.FiniteStrainBehaviourOptions()
bopts.stress_measure = mgis_bv.FiniteStrainBehaviourOptionsStressMeasure.PK2
bopts.tangent_operator = (
mgis_bv.FiniteStrainBehaviourOptionsTangentOperator.DPK1_DF
)
behaviour = mgis_bv.load(bopts, lib_path, behaviour_name, hypothesis)
else:
print("Small strain behaviour detected.")
behaviour = mgis_bv.load(lib_path, behaviour_name, hypothesis)Small strain behaviour detected.The Behaviour object now contains several information
about the behaviour implementation such as names and types of gradients
and thermodynamic forces and external or internal state variables.
Here, we work with a purely mechanical behaviour so that gradients
consist only of the total strain Strain. The resulting
thermodynamic stress is Stress.
By default, Temperature is always declared as an
external state variable by MFront.
Note
The automatic declaration of the
Temperaturecomes from the fact that many solvers adopted theUMATinterface which treats the temperature specifically.If portability is not an issue (the
genericinterface is the only one that supports this feature), the automatic declaration can be disabled by modifying the declaration of the domain specific language in theMFrontfile as follows:@DSL Implicit{ automatic_declaration_of_the_temperature_as_first_external_state_variable: false }
Finally, since we are dealing with an elastoplastic behaviour with
isotropic hardening, the internal state variables are the
ElasticStrain and the EquivalentPlasticStrain.
The former is a symmetric tensor whereas the latter is only a
scalar.
print("Kinematic type:", behaviour.getKinematic())
print("Gradient names:", [s.name for s in behaviour.gradients])
print("Thermodynamic forces names:", [s.name for s in behaviour.thermodynamic_forces])
print(
"External state variable names:",
[s.name for s in behaviour.external_state_variables],
)
print(
"Internal state variable names:",
[s.name for s in behaviour.internal_state_variables],
)
print(
"Internal state variable types:",
[s.type for s in behaviour.internal_state_variables],
)Kinematic type: SmallStrainKinematic
Gradient names: ['Strain']
Thermodynamic forces names: ['Stress']
External state variable names: ['Temperature']
Internal state variable names: ['ElasticStrain', 'EquivalentPlasticStrain']
Internal state variable types: [mgis.behaviour.VariableType.STENSOR, mgis.behaviour.VariableType.SCALAR]The size of these different quantities can be checked. Symmetric tensors in 3D are represented here as arrays of size 6 (using Mandel notation, see Tensors in MFront).
for s in behaviour.gradients + behaviour.thermodynamic_forces + behaviour.internal_state_variables:
print(f"{s.name} is of size {mgis_bv.getVariableSize(s, hypothesis)}")Strain is of size 6
Stress is of size 6
ElasticStrain is of size 6
EquivalentPlasticStrain is of size 1For more complex behaviours, different “blocks” of tangent operators can be declared and computed. The names of the tangent blocks can be retrieved as shown below. Here, we have a simple mechanical behaviour so that the only tangent operator is the tangent stiffness \(\boldsymbol{C}_\text{tang} = \dfrac{\partial \boldsymbol{\sigma}}{\partial \boldsymbol{\varepsilon}}\).
print(
"Tangent operator block names:",
[f"d{s[0].name}_d{s[1].name}" for s in behaviour.tangent_operator_blocks],
)Tangent operator block names: ['dStress_dStrain']Finally, names of material properties can also be retrieved easily as follows:
print(
"Material property names:",
[s.name for s in behaviour.material_properties],
)Material property names: ['InitialYieldStress', 'FinalYieldStress', 'HardeningCoefficient']Other information such as behaviour material symmetry or predefined parameters can also be obtained as follows:
print(behaviour.symmetry)
print(behaviour.parameters)Isotropic
['epsilon', 'theta', 'YoungModulus', 'PoissonRatio', 'RelativeValueForTheEquivalentStressLowerBoundDefinition', 'minimal_time_step_scaling_factor', 'maximal_time_step_scaling_factor', 'numerical_jacobian_epsilon']The material data manager will be the data structure which will hold the information about the mechanical state and tangent operator for a single or a batch of integration points during a given time step. We first set up the data manager by declaring the number of integration points we want to handle.
ngauss = 3
data = mgis_bv.MaterialDataManager(behaviour, ngauss)Values at the beginning of the time step will be stored in
data.s0. This object is a StateManager which
holds values of the gradients, thermodynamic forces and internal state
variables.
A similar one is stored in data.s1 which will hold
values at the end of the current time step, once behaviour integration
has been performed.
Here, by default, Strain is initialized with a zero
tensor, as well as all other variables. We see below that
data.s0.gradients is of shape ngauss x 6
corresponding to the 6 components of the ngauss integration
points.
print("Gradients:")
print(" Beginning of time step:\n", data.s0.gradients)
print(" End of time step:\n", data.s1.gradients)
print("Thermodynamic forces:")
print(" Beginning of time step:\n", data.s0.thermodynamic_forces)
print(" End of time step:\n", data.s1.thermodynamic_forces)Gradients:
Beginning of time step:
[[0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0.]]
End of time step:
[[0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0.]]
Thermodynamic forces:
Beginning of time step:
[[0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0.]]
End of time step:
[[0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0.]]Conversely, internal state variables are of shape
ngauss x 7 with components of ElasticStrain
and EquivalentPlasticStrain being concatenated.
print("Internal state variables:")
print(" Beginning of time step:\n", data.s0.internal_state_variables)
print(" End of time step:\n", data.s1.internal_state_variables)Internal state variables:
Beginning of time step:
[[0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0.]]
End of time step:
[[0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0.]]In order to perform the behaviour integration, we need to declare the values for the material properties which are yet unspecified. We also need to declare the value of the temperature, although it is not used in practice. Note that we can declare different values at the beginning and at the end of the time step, although here we use the same values. Finally, we would also apply different values for the different integration points, if needed.
for state_manager in [data.s0, data.s1]:
mgis_bv.setMaterialProperty(state_manager, "InitialYieldStress", 250e6)
mgis_bv.setMaterialProperty(state_manager, "FinalYieldStress", 400e6)
mgis_bv.setMaterialProperty(state_manager, "HardeningCoefficient", 1000.)
mgis_bv.setExternalStateVariable(state_manager, "Temperature", 293.15)Below, we will impose a cyclic loading in terms of imposed horizontal
strain. The value of the time step dt must be provided but
is irrelevant here since we are dealing with a rate-independent
behaviour.
# strain history
N = 20
eps_max = 3e-3
eps_list = np.concatenate((np.linspace(0, eps_max, N), np.linspace(eps_max, -eps_max, N), np.linspace(-eps_max, 0, N))*2)
dt = 0We now implement the expression of the global residual \(\boldsymbol{R}\) and its jacobian \(\boldsymbol{J}\). As stresses are expressed
here in Pa, we use a scaling of 1e-9 to scale the stress
components to values close to strain values.
stress_scaling = 1e-9
def residual(eps, sig, imposed_eps):
return np.concatenate((eps[:,[0]]-imposed_eps, sig[:,1:]*stress_scaling), axis=1)
def jacobian(eps, sig, Ct):
Jac = np.zeros_like(Ct)
Jac[:,0,0] = 1
Jac[:,1:,:] = Ct[:,1:,:]*stress_scaling
return JacIntegration of the material behaviour will be done by specifying that we want to compute the consistent tangent operator.
We then implement the time stepping loop and the Newton-Raphson resolution.
We recall that the values of the total strain are all assumed to be
unknown and will be stored in the eps variable, the latter
being initialized with the strain values at the beginning of the time
step.
During a Newton-Raphson iteration, the strain values at the end of
the time step are updated with the current value for eps.
Then, integration is performed using mgis_bv.integrate.
Next, we retrieve the new value for the stress, stored in
sig and of the tangent operator, stored in
Ct.
We evaluate the residual \(\boldsymbol{R}\) and perform the Newton
correction to eps by solving the jacobian system. Finally,
we report the horizontal stress value at the first integration point (we
can check that all ngauss points give the same value).
Before going to the next load step, the computed state must be
updated using mgis_bv.update(data) which copies the
computed state s1 into s0.
integration_type = mgis_bv.IntegrationType.IntegrationWithConsistentTangentOperator
rtol = 1e-8
atol = 1e-8
niter_max = 20
Sxx = np.zeros_like(eps_list)
for i, eps_imposed in enumerate(eps_list):
eps = np.copy(data.s0.gradients)
sig = np.copy(data.s0.thermodynamic_forces)
# Newton-Raphson solve
nres = 1
nres0 = max(np.linalg.norm(residual(eps, sig, eps_imposed),axis=1))
niter = 0
while (nres > max(atol, rtol * nres0)) and (niter < niter_max):
data.s1.gradients[:] = eps
integration_status = mgis_bv.integrate(data, integration_type, dt, 0, ngauss)
assert integration_status > 0, "Behaviour integration has failed."
sig = data.s1.thermodynamic_forces
Ct = data.K
res = residual(eps, sig, eps_imposed)
nres = max(np.linalg.norm(res, axis=1))
# print(f"Iteration {niter}, residual = {nres}")
Jac = jacobian(eps, sig, Ct)
for k in range(ngauss):
eps[k,:] += np.linalg.solve(Jac[k,:,:], -res[k,:])
niter += 1
assert niter <= niter_max, "Newton method did not converge within maximum number of iterations."
Sxx[i] = data.s1.thermodynamic_forces[0, 0]
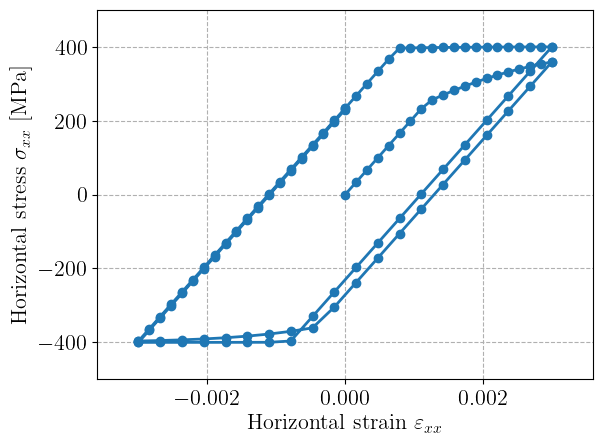
mgis_bv.update(data)Plotting the horizontal stress strain evolutions shows the expected behaviour with a first yielding at 250 MPa and a saturation at 400 MPa in both tension and compression.
import matplotlib.pyplot as plt
plt.plot(eps_list, Sxx*1e-6, '-o')
plt.xlabel(r"Horizontal strain $\varepsilon_{xx}$")
plt.ylabel(r"Horizontal stress $\sigma_{xx}$ [MPa]")
plt.xlim(-1.2*eps_max, 1.2*eps_max)
plt.ylim(-500, 500)
plt.show()